About

AI scientist with over 5 years of research experience applying machine learning to solve problems in protein design. Experienced in working across interdisciplinary teams and building user-driven scientific solutions. Seasoned software developer proficient in Python and C/C++, with experience in designing, training, and fine-tuning large language models for biology.
Publications
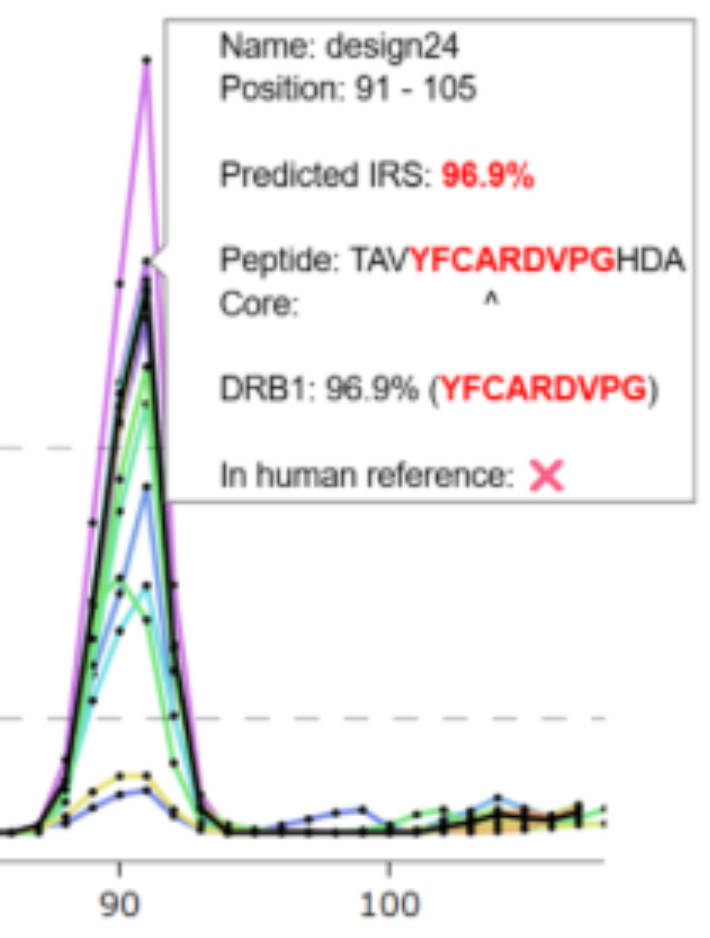
ImmunoGeNN: Accelerating Early Immunogenicity Assessment for Generative Design of Biologics
MH Høie, B Reynisson, P Marcatili, J Ferkinghoff-Borg, K Lamberth, KL Kopp, M Nielsen, VI Jurtz
EurIPS SIMBIOCHEM, 2025
PDF
Abstract
Bibtex
Webserver

AntiFold: improved structure-based antibody design using inverse folding
MH Høie, A Hummer, TH Olsen, M Nielsen, C Deane
Bioinformatics Advances, 2025
PDF
Abstract
Bibtex
Webserver
PhD thesis - AI in Antibody Design
MH Høie
DTU Orbit, 2024
PDF
Abstract
Bibtex
Comparative Structure Analysis of the Multi-Domain, Cell Envelope Proteases of Lactic Acid Bacteria
LF Christensen, MH Høie, CH Bang-Berthelsen, P Marcatili, EB Hansen
MDPI Microorganisms, 2023
PDF
Abstract
Bibtex
DiscoTope-3.0 - Improved B-cell epitope prediction using inverse folding latent representations
MH Høie, FS Gade, JM Johansen, C Würtzen, O Winther, M Nielsen, P Marcatili
Frontiers in Immunology, 2024
PDF
Abstract
Bibtex
Webserver
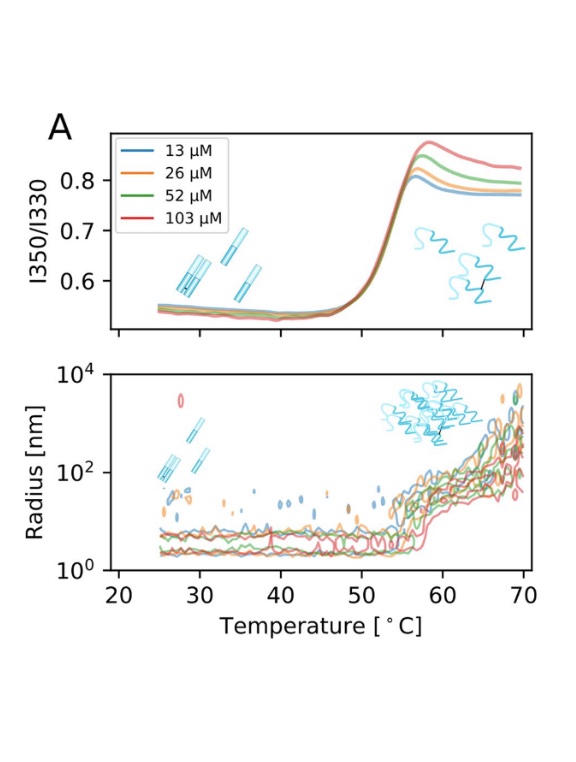
Widespread amyloidogenicity potential of multiple myeloma patient-derived immunoglobulin light chains
R Sternke-Hoffman, T Pauly, RK Norrild, J Hansen, F Tucholski, MH Høie, P Marcatili, M Dupré, M Duchateau, M Rey, C Malosse, S Metzger, A Boquoi, F Platten, SU Egelhaaf, J Chamot-Rooke, R Fenk, L Nagel-Steger, R Haas, AK Buell
BMC Biology, 2023
PDF
Abstract
Bibtex
BepiPred-3.0 - Improved B-cell epitope prediction using protein language models
J Clifford, MH Høie, S Deleuran, B Peters, M Nielsen, P Marcatili
Protein Science, 2022
PDF
Abstract
Bibtex
Webserver
NetSurfP-3.0 - accurate and fast prediction of protein structural features by protein language models and deep learning
MH Høie, EN Kiehl, B Petersen, M Nielsen, O Winther, H Nielsen, J Hallgren, P Marcatili
Nucleic Acids Research, 2022
PDF
Abstract
Bibtex
Webserver
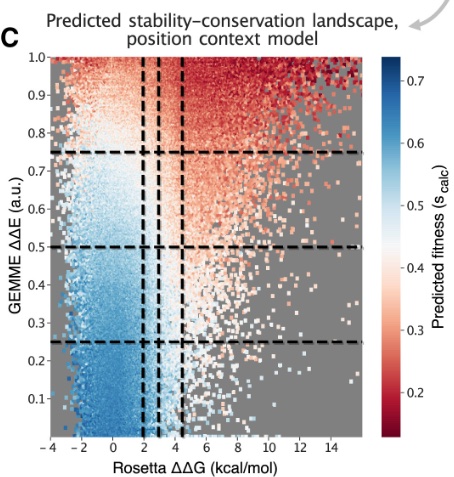
Predicting and interpreting large-scale mutagenesis data using analyses of protein stability and conservation
MH Høie, M Cagiada, AHB Frederiksen, A Stein, K Lindorff-Larsen
Cell Reports, 2022
PDF
Abstract
Bibtex
Website
Talks & workshops
Talks and workshops at selected conferences.
ImmunoGeNN: Proteome-scale screening of MHC-II immunogenicity risk in the global population
Annual Danish Bioinformatics Conference 2025
October 2025
De novo generation of antibody binders with RFantibody
Workshop, BioIT Boston 2025.
Click here for Jupyter notebook
April 2025
AntiFold: Improved antibody structure design using inverse folding
Spotlight, NeurIPS GenBio 2023
Dec 2023
DiscoTope-3.0: Improved B-cell epitope prediction using AlphaFold2 modeling and inverse folding latent representations
Selected talk, ELIXIR Annual Danish Bioinformatics Conference 2023
Sep 2023
Immunological binding prediction using protein language models & inverse folding
SDC Annual Life Science Engineering and Informatics PhD Symposium 2023
June 2023
Supervising
Below are listed titles of the M.Sc. Thesis' and independent research projects I have co-supervised during my time at the Technical University of Denmark.
Early MHC class I prediction for immunogenicity of gene editing protein product therapies
Master Thesis, 30 ECTS
Lorenzo Favaro
Spring 2025
Unsupervised contrastive learning for antibody structure clustering
Master Thesis, 30 ECTS
Christian Peder Jacobsen
Autumn 2023
Development of improved prediction methods for B cell epitopes prediction
Special project, 5 ECTS
Carlos de Santiago León
Spring 2023
Structure-based prediction of TCR-pMHC interaction using Graph Neural Networks
Master Thesis, 30 ECTS
Julie Maria Johansen,
Charlotte Würtzen
Autumn 2022
BepiPred-3.0: Improved B-cell epitope prediction using protein language models
Master Thesis, 30 ECTS
Joakim Clifford
Autumn 2021
Using deep learning for improving TCR homology modeling and its application to immunogenicity prediction
Master Thesis, 30 ECTS
Ida Meitil
Spring 2021
Community
To build a community in Denmark for sharing experience on working with machine-learning and data-science methods handling biological data, I organized events together with Paolo Marcatili, Tobias Hegelund Olsen and Andreas Fønss Møller. This led to 4 events with 200+ academic and industry professional participants, attending from Novo Nordisk, NovoZymes, Lundbeck, DTU and KU.
Biodatascience101
Organized 4 events teaching biological data-science to academics and industry professionals
2019-2021
Website

Copenhagen Bioinformatics Hackathon 2021
Hosted TCR-p-MHC prediction challenge, with 7 competing teams
Spring 2021
Website
Teaching
Below you can find a list over courses I have taught in.
DTU course 22117 Protein structure and computational biology
Teaching assistant
Spring 2022

DTU course 22111 Introduction to Bioinformatics
Teaching assistant
Spring 2022

DTU course 22110 Python and Unix for Bioinformaticians
Teaching assistant
Autumn 2021
Book chapters
Scientific book chapters in collaboration with others.
In Silico Tools for Predicting Novel Epitopes
C Barra, JB Nilsson, A Saksager, I Carri, S Deleuran, HMG Alvarez, MH Høie, Y Li, JN Clifford, YTR Wan, LS Moreta, M Nielsen
Intracellular Pathogens, August 2024
Media
Selected articles on my research.
Dynamo magazine nr. 78
September 2024

DTU Career as a PhD student
August 2024